squigualiser
Base shift calculation
- User can shift the base sequence to the left by
nnumber of bases by providing the argument--base_shift -ntoplotandplot_pileupcommands. - This is helpful to correct the signal level to the base. A negative
nvalue will shift the base sequence to the left. - Base shift concept was motivated from the idea presented in pore_model document about the most contributing base to the current level.
- We can programmatically calculate a
base_shiftto nicely align the signal to the base (color). The calculation is implemented here. - However, the user is adviced to use
--profile(documented here) which automatically sets the--base_shift.
Example 1
- At site
A21there is a heterozygous variantA/T. - The first pileup (Fig. 1) has a wrong base shift (-5) and second pileup (Fig. 2) has the correct base shift (-6).
- Hence, the second pileup (Fig. 2) has both the variant (blue box - bed annotation) and the signal differences aligned to each other.
- If the base shift was some other value instead of
-6the difference in the signals will not align with base. -
This variant example is documented here.
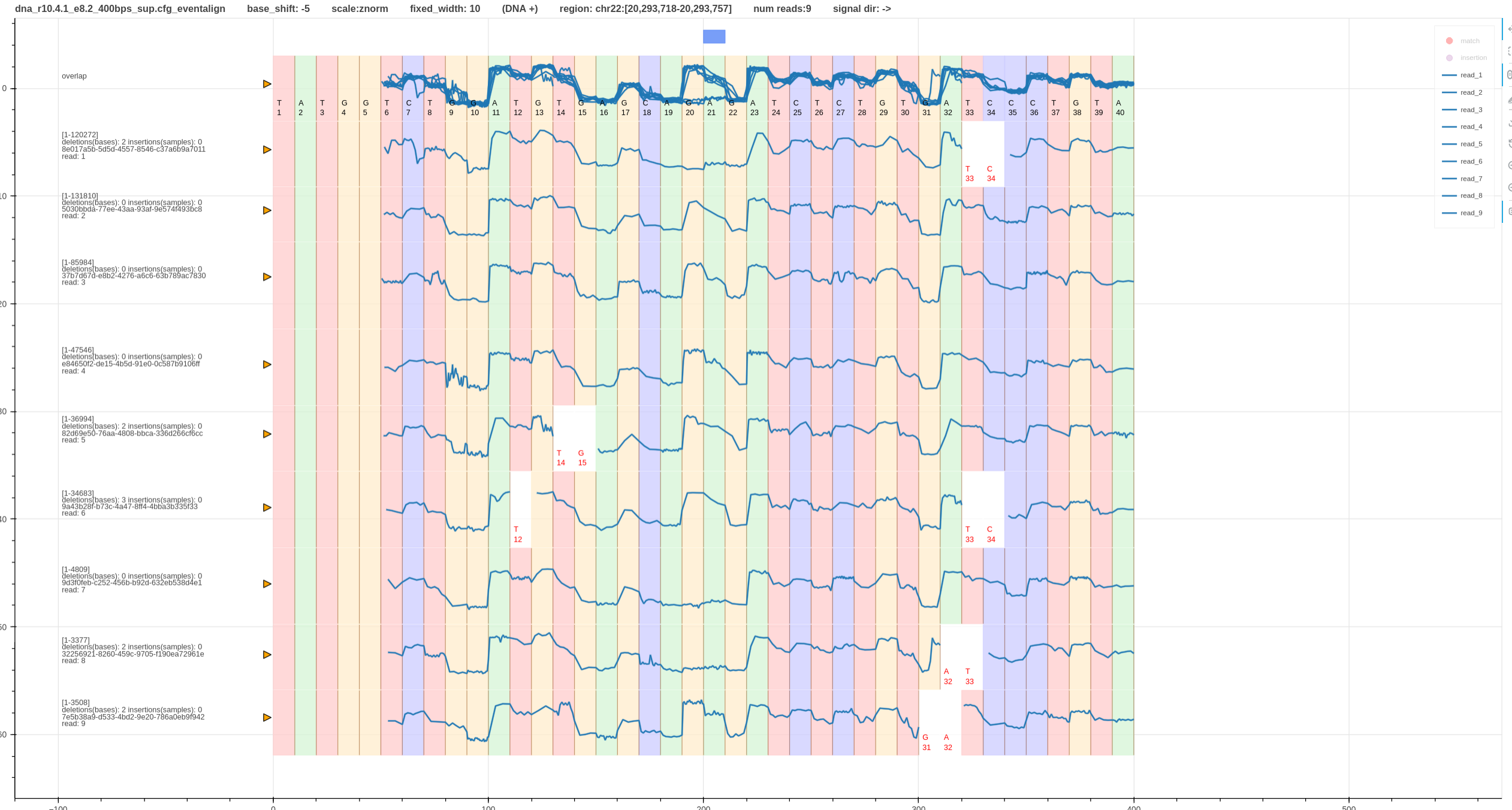 Figure 1
Figure 1 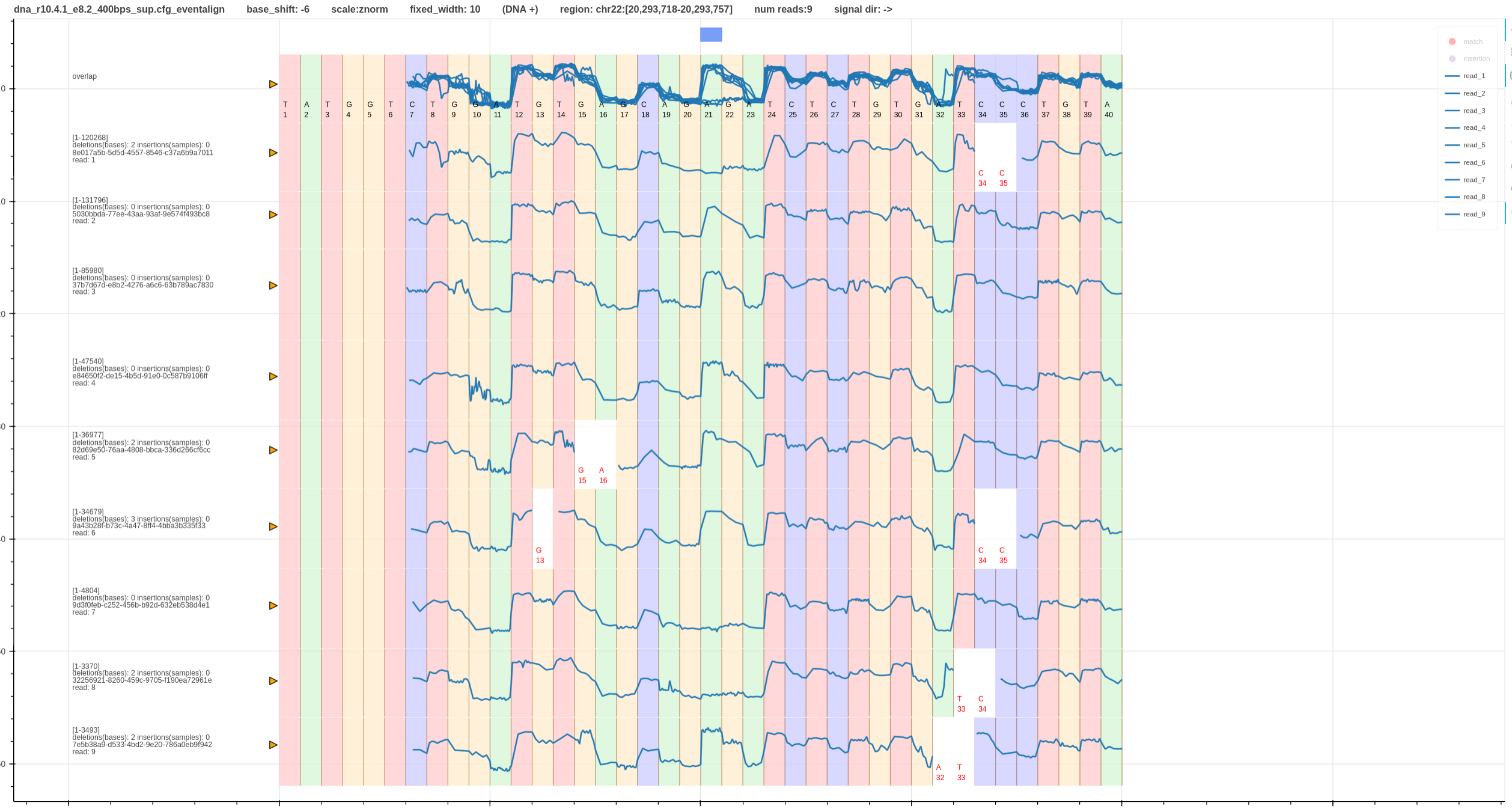 Figure 2
Figure 2
Example 2
- Fig. 3 and 4 have the same
dna_r10.4.1_e8.2_400bpssignal pileup with a base shift of0and-6respectively. - The base colors in Fig. 3 do not nicely aligned to the signal.
- However, in Fig. 4 the signal is moving from low to high when a base
Tis met. - These signal pileups were generated using f5c eventalign.
- F5c used
dna_r10.4.1_e8.2_400bpspore model to align these signals. Its most contributing base index is-6and hence the appropriate base shift in this scenario is also-6.
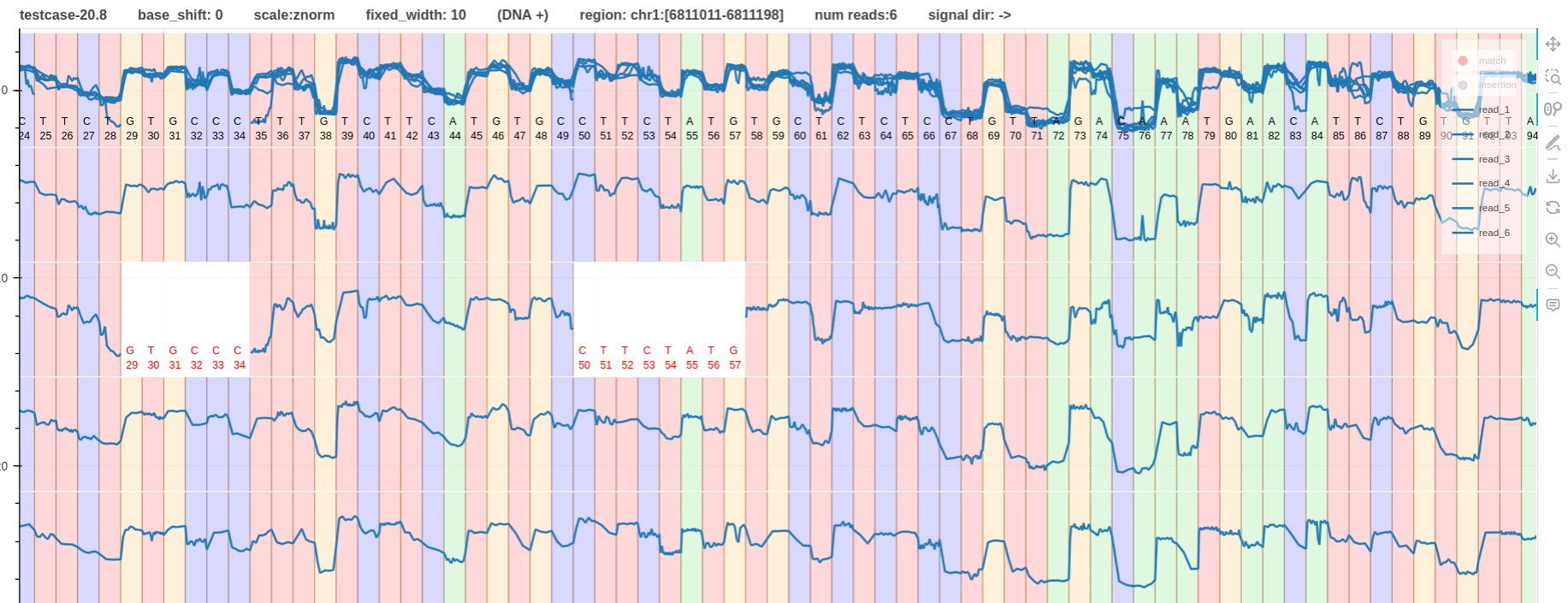
Figure 3: base shift 0 link
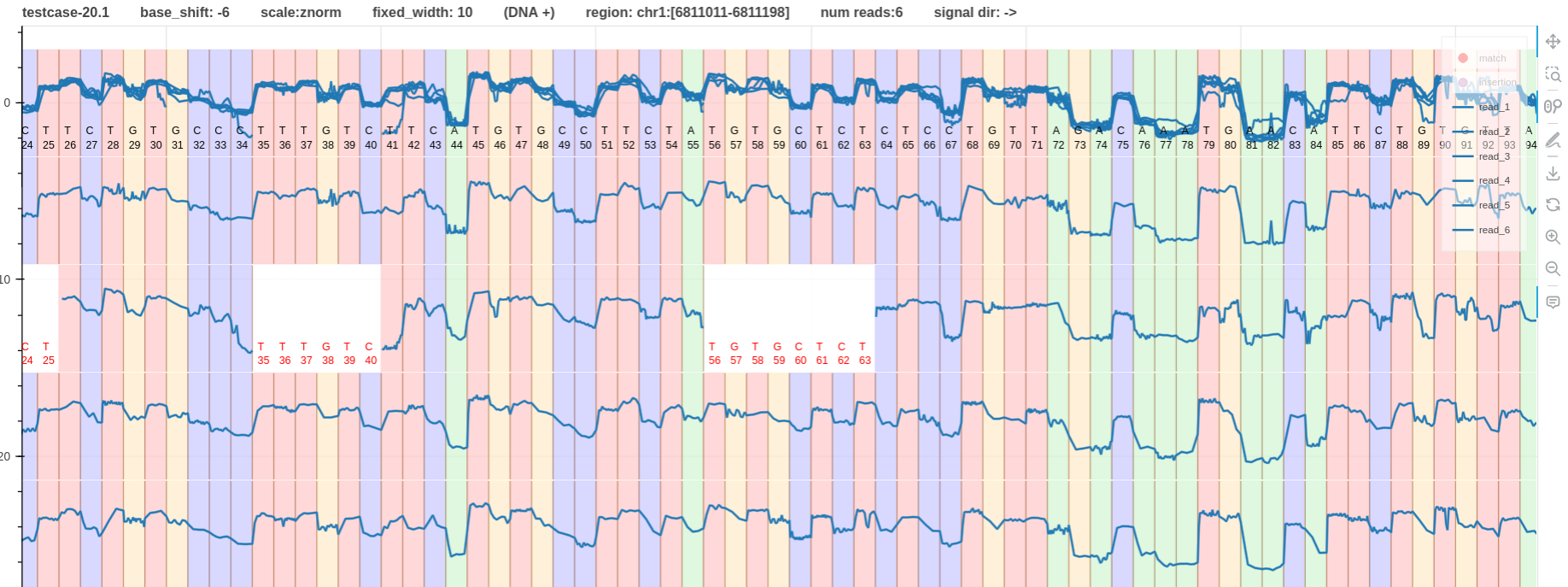
Figure 4: base shift -6 link
Example 3
- Consider a
dna_r10.4.1_e8.2_400bpsforward and reverse mapped pileups for the same genomic region. - Fig.5 has
0base shift for both tracks. - In Fig. 6, note that the reverse mapped pileup has a
-2base shift. This is because the signal sequencing direction is from right to left (more information). - In Fig. 6, both pileups have the signals going from low to high when a base
Tis met.
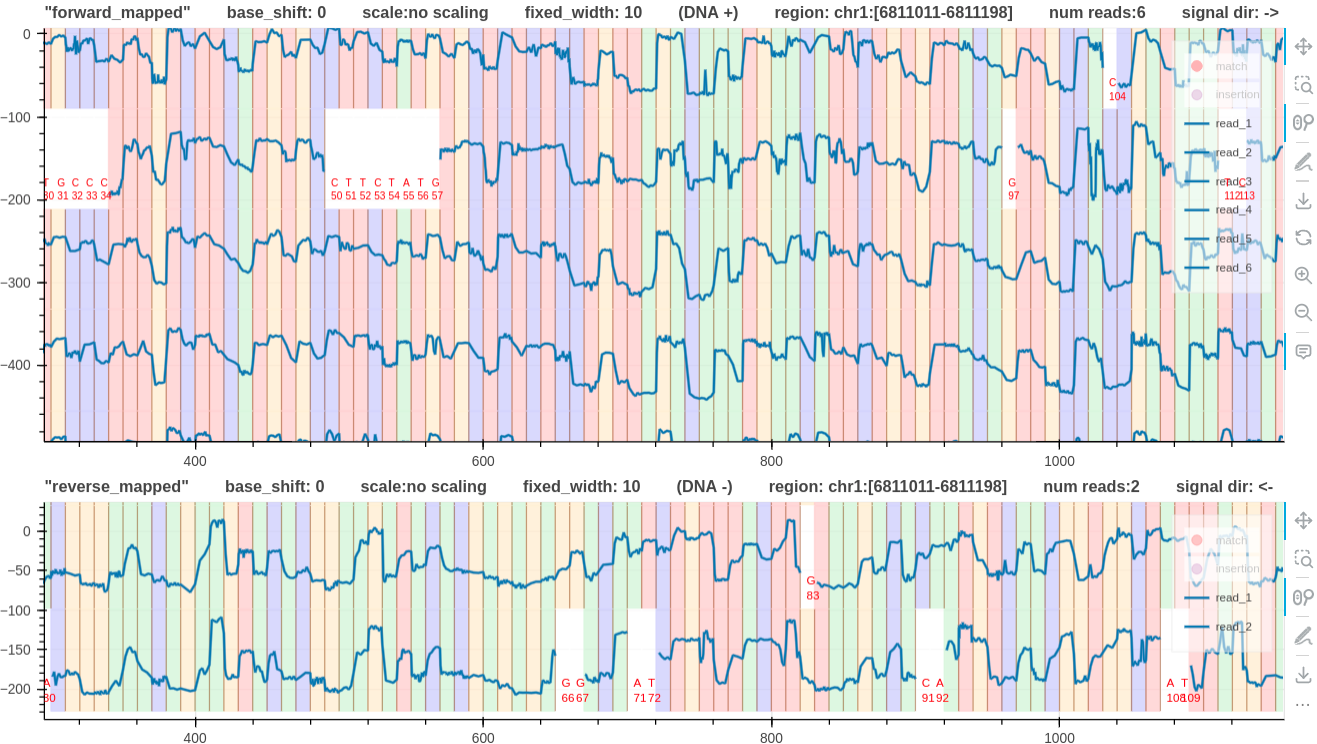
Figure 5: base shift 0, 0 link
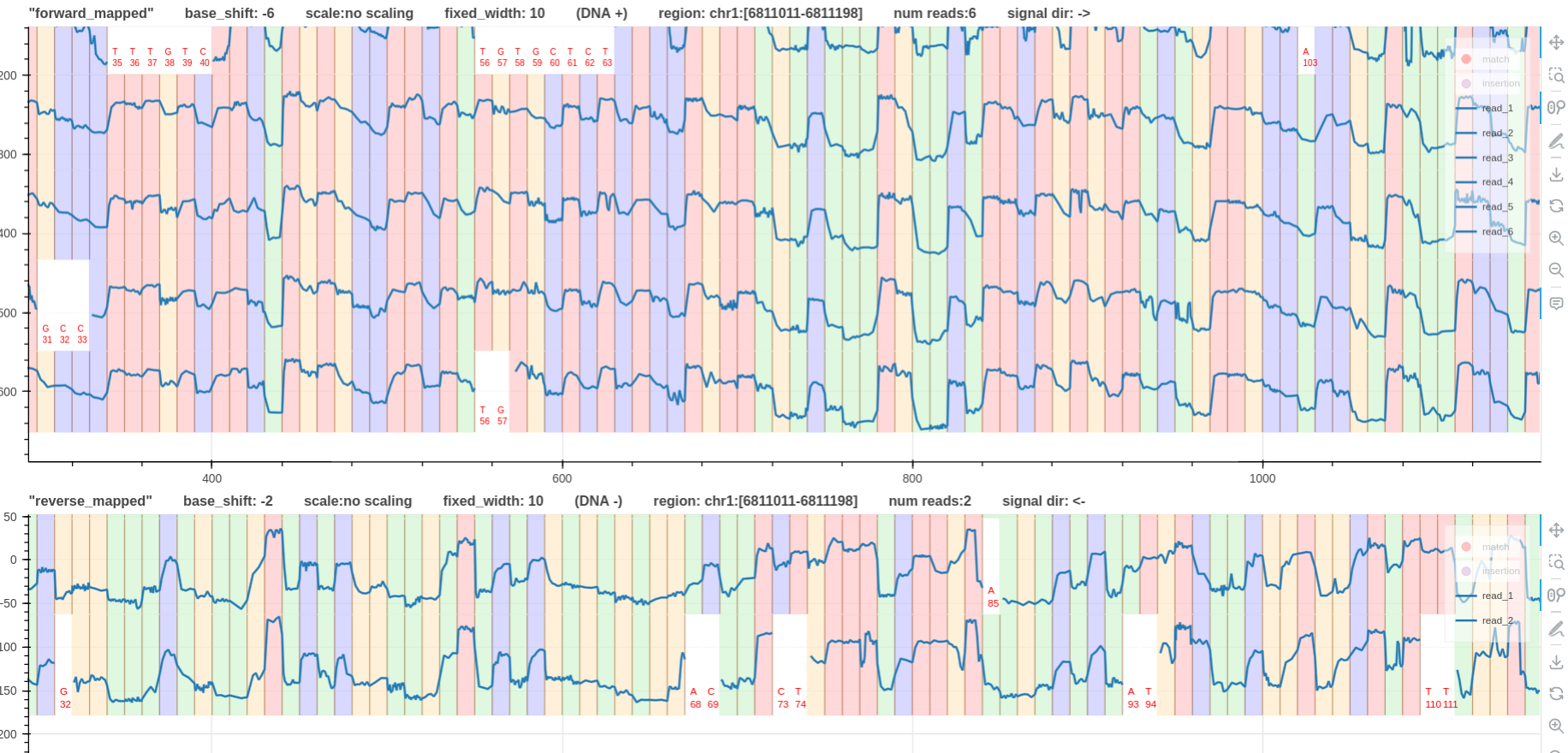
Figure 6: base shift -6, -2 link
Example 4
Fig. 7 shows the forward and reverse mapped pileups generated using f5c eventalign for dna_r9.4.1_450bps data. F5c used the 6mer model (more information).
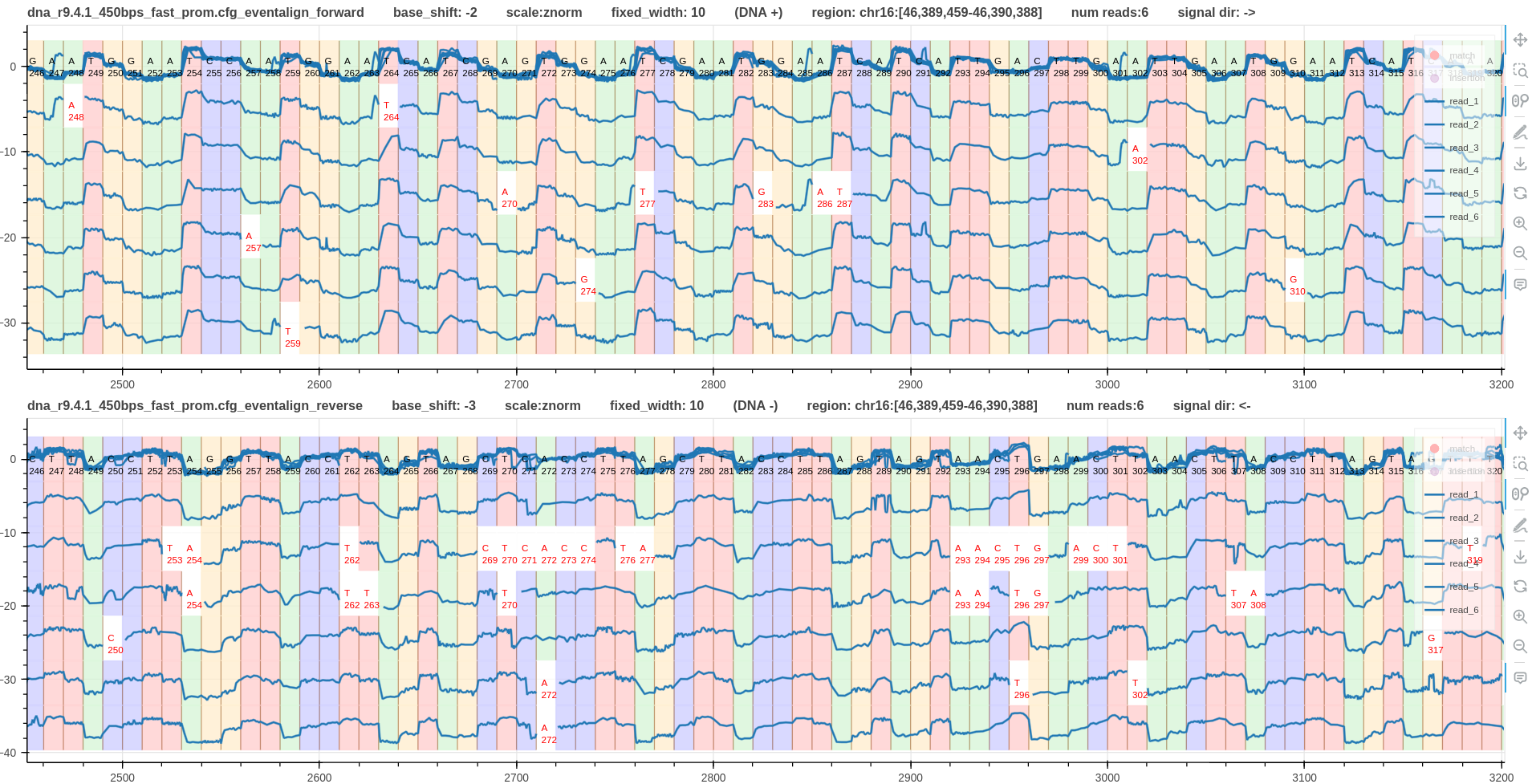
Figure 7: base shift -2, -3
Example 5
- Fig. 8 shows the forward mapped pileups generated using f5c eventalign for
rna_r9.4.1_70bpsdata. F5c used the rna 5mer model (more information). - Note that
squigualiseralways plot the RNA reads in its correct sequencing direction (reverse mapped RNA reads are skipped; reverse mapped RNA reads exist if a genome was used as the reference instead of a transcriptome).
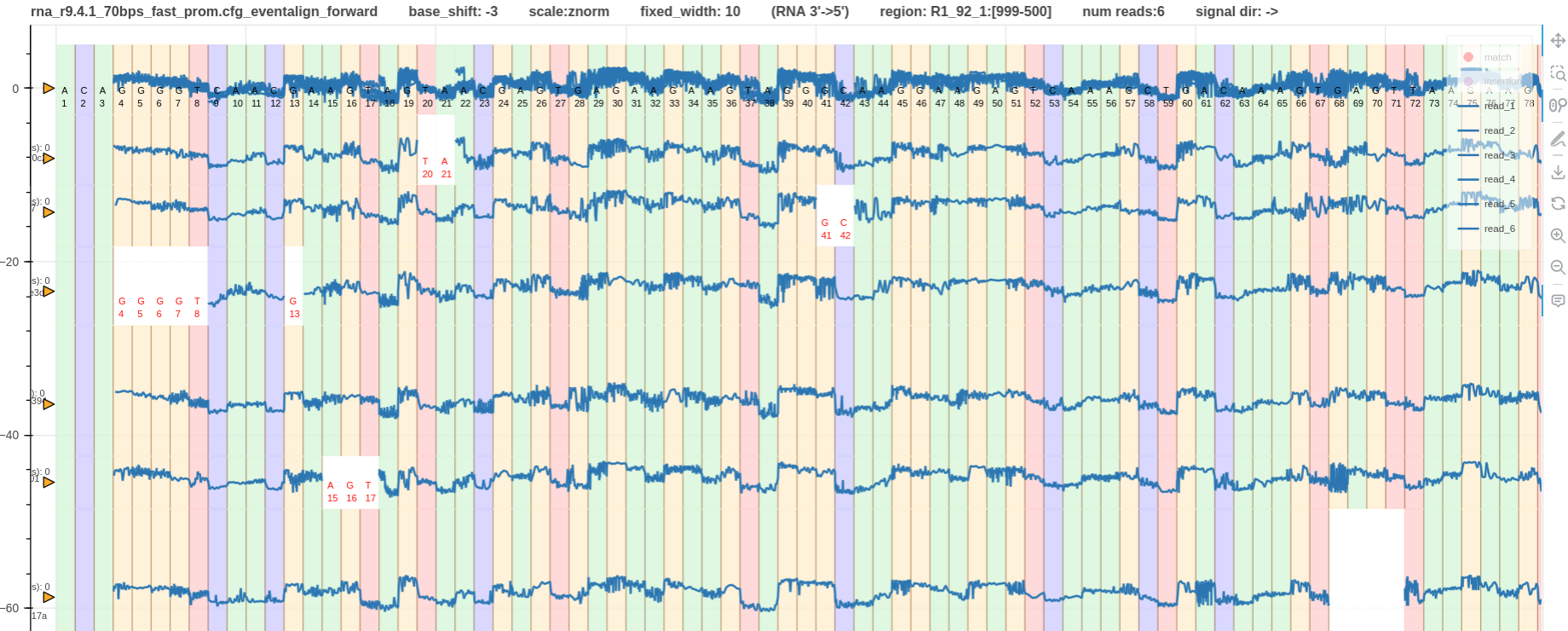
Figure 8: base shift -3