squigualiser
Pipeline methylation detection DNA
In this example three cpg methylations are found in humangenome hg38.fa in the region chr1:92,779,392-92,779,445.
This is the same region that we used in the variant calling pipeline
Fig. 1 shows a screenshot of IGV.
The first track is the bam file containing alignment track.
The second track is the high confidence variants vcf file (subset.vcf.gz).
The third track is the methylated cpg sites based on f5c methylation-call tool (meth_freq.bigwig).
The last track shows the aligned reads.
Methylated cites are present at chr1:92,779,423, chr1:92,779,428, and chr1:92,779,436.
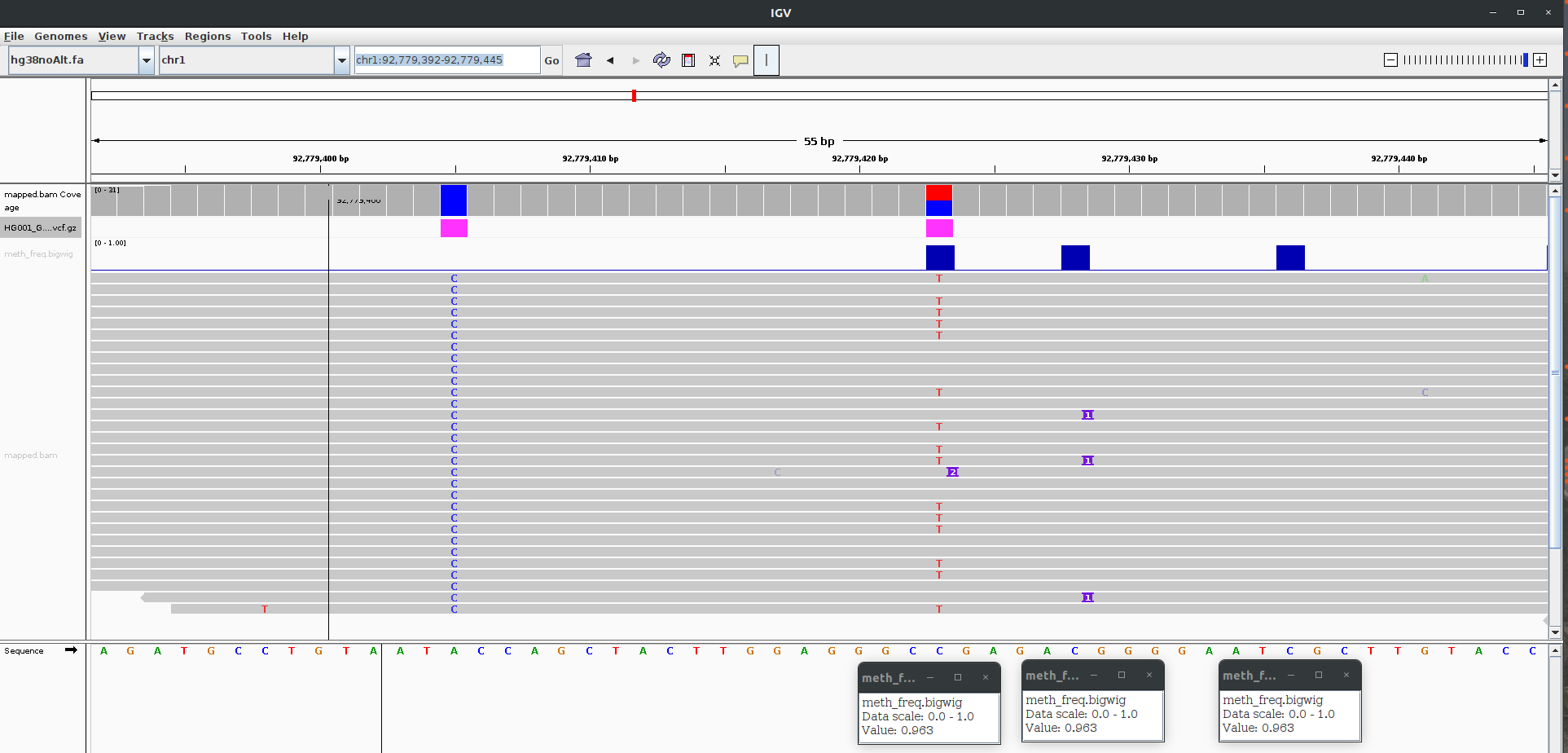
Figure 1
The pipeline data is at test/data/raw/pipelines/pipeline_2/dna_r10.4.1_e8.2_400bps.
Note that the pipeline must be executed inside the virtual environment where squigualiser is installed.
The bash script run.sh is very similar to the pipeline explained here.
Fig.2 shows two tracks. The first is the reference to signal alignment using eventalign.
The second is the reference simulated signal.
The blue color boxes in the first track annotate the methylated cites as shown on IGV track.
The real signal plot clearly shows two different current levels at methylated cites.
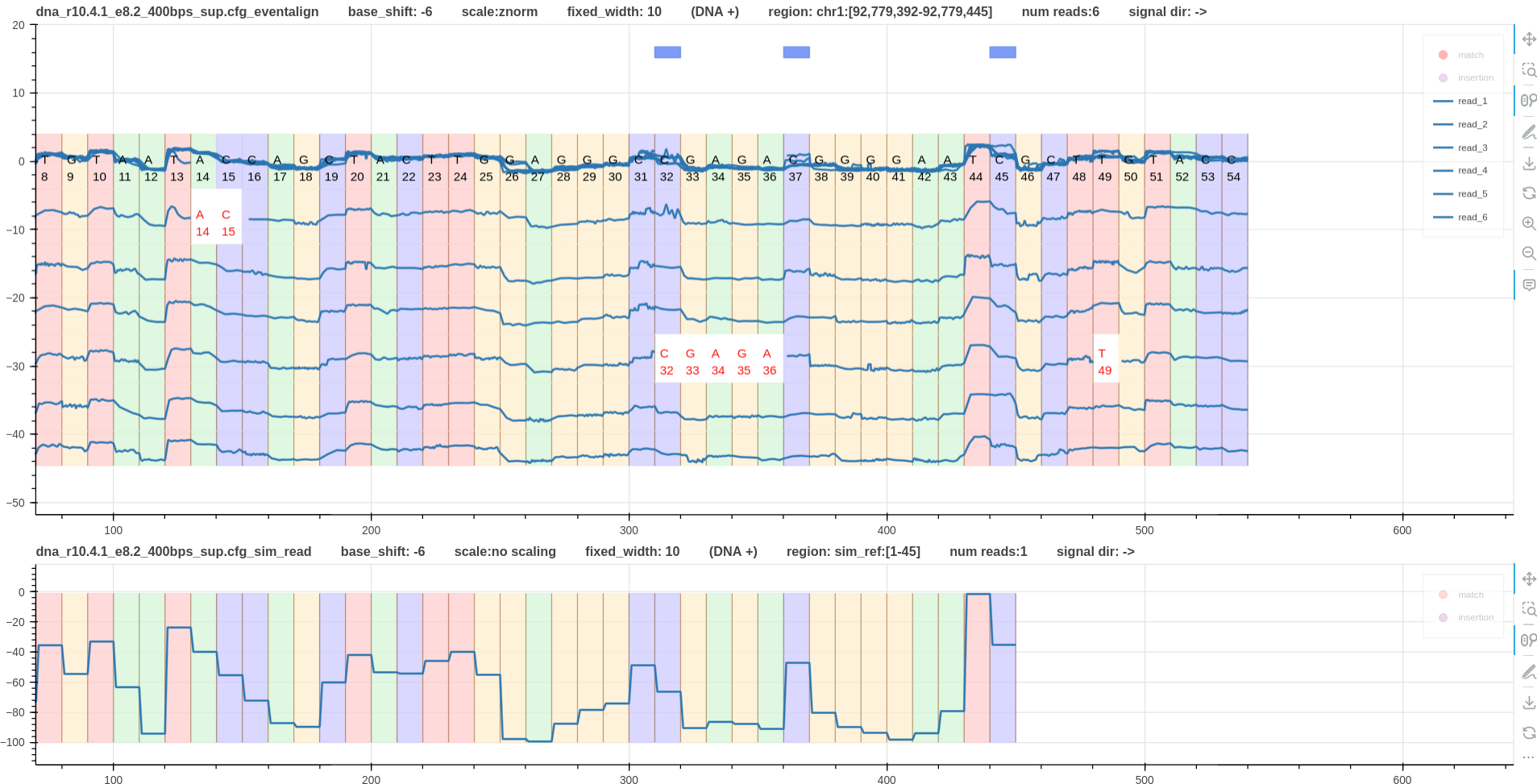
Figure 2
Using methylation frequency information
Optionally, we can use the methylation frequency information to better visualise the methylations.
- Filter a site from the methylation frequency
meth-freq.tsvfile where the frequency is approximately0.5. - Filter the
+reads of the filtered site from the rawmeth.tsvfile. - Filter the read ids where the log likelihood ratio is less than
-2and above2for unmethylated and methylated reads respectively. - Use the filtered read ids to create a pileup plot.
In the following example we considered the + strand reads covering the site chr1:92790686 with a methylation frequency of 0.542.
As shown in the overlap plot in Fig.3 the site C18 has two distinct current levels.
The higher current level corresponds with the unmethylated sites/read_ids.
The lower current level corresponds with the methylated sites/read_ids.
Out of the 11 total reads in the pileup, 3 have deletions at the site, 3 are unmethylated, and 5 are methylated (Table 1).
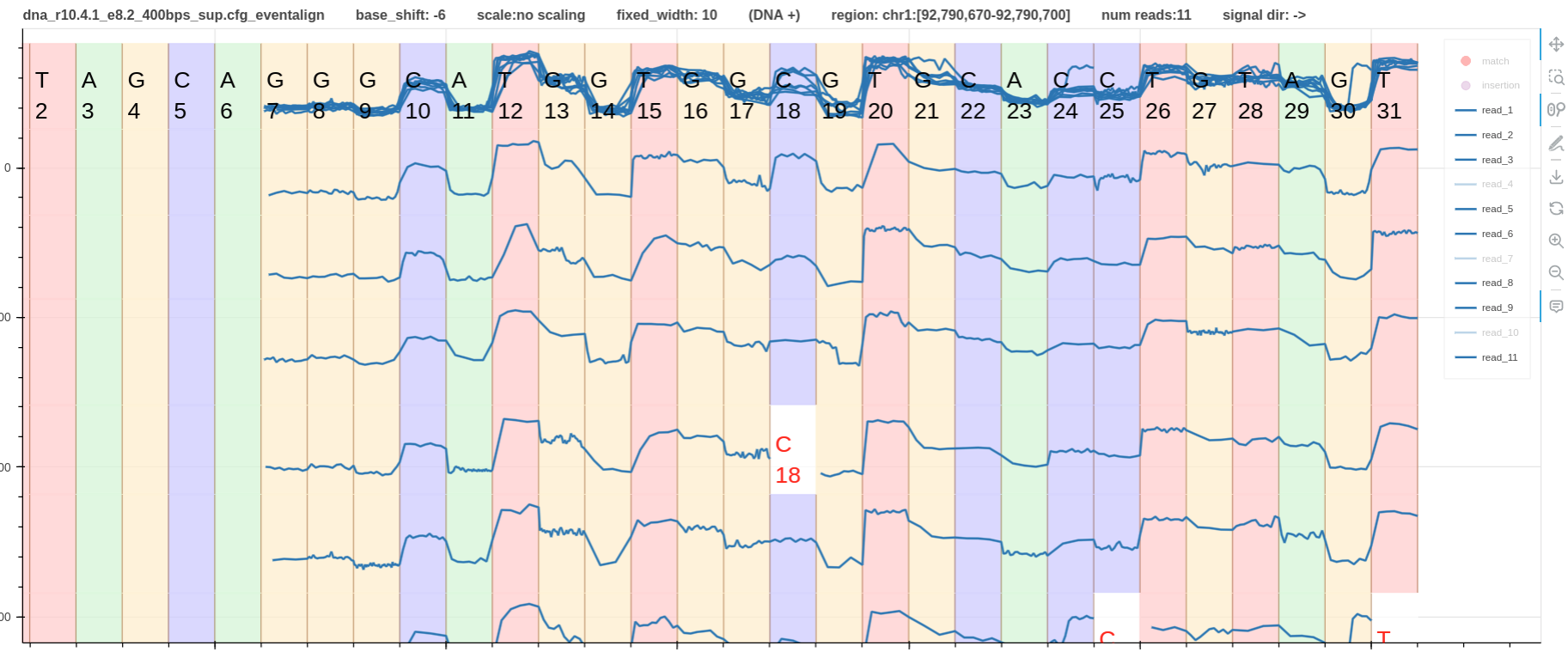
Figure 3
Table 1
| chromosome | strand | start | end | read_name | log_lik_ratio | log_lik_methylated | log_lik_unmethylated | num_calling_strands | num_motifs |
|---|---|---|---|---|---|---|---|---|---|
| chr1 | + | 92790686 | 92790686 | 1e12424f-dcab-4cd0-83c3-79afc1326199 | -4.41 | -120.27 | -115.86 | 1 | 1 |
| chr1 | + | 92790686 | 92790686 | 6b960395-8f61-4b8f-880d-e1852a5556a8 | -2.78 | -147.48 | -144.7 | 1 | 1 |
| chr1 | + | 92790686 | 92790686 | d2d2c018-f49f-436b-a038-e441f95a49ad | -2.69 | -162.08 | -159.39 | 1 | 1 |
| chr1 | + | 92790686 | 92790686 | fdb21051-2388-446d-acf2-a6848669ced8 | 2.62 | -84.27 | -86.89 | 1 | 1 |
| chr1 | + | 92790686 | 92790686 | caad6a89-8ad5-485d-9ed9-903e99ed1b6a | 3.8 | -196.16 | -199.95 | 1 | 1 |
| chr1 | + | 92790686 | 92790686 | 362dc957-024e-4c56-a26a-c7fe4cd07304 | 4.15 | -198.4 | -202.56 | 1 | 1 |
| chr1 | + | 92790686 | 92790686 | 5ae057b3-cde0-4248-a905-d18a75079ba2 | 4.6 | -127.22 | -131.82 | 1 | 1 |
| chr1 | + | 92790686 | 92790686 | 0aa2c317-d8c4-4861-ac6f-87bab7fff2d8 | 4.77 | -134.04 | -138.81 | 1 | 1 |
| chr1 | + | 92790686 | 92790686 | b23eadb5-9f1f-454b-82c9-1d86b0d6faf8 | 5.39 | -106.46 | -111.84 | 1 | 1 |
| chr1 | + | 92790686 | 92790686 | 9efe2d41-af4e-4b6c-9f20-48e971ac06b3 | 5.54 | -117.49 | -123.03 | 1 | 1 |
| chr1 | + | 92790686 | 92790686 | 97953cda-3ab0-4f1f-9e6a-47a04e75bf0a | 5.86 | -125.13 | -130.99 | 1 | 1 |
The plot is available here.