squigualiser
Move table
Nanopore basecallers output move arrays in SAM/BAM format. The important fields are listed below.
- read_id
- basecalled fastq sequence length
- basecalled fastq sequence
- raw signal length in
nstag - raw signal trim offset in
tstag - move table in
mvtag - stride used in the neural network (down sampling factor) in
mvtag
An example move table looks like the following,
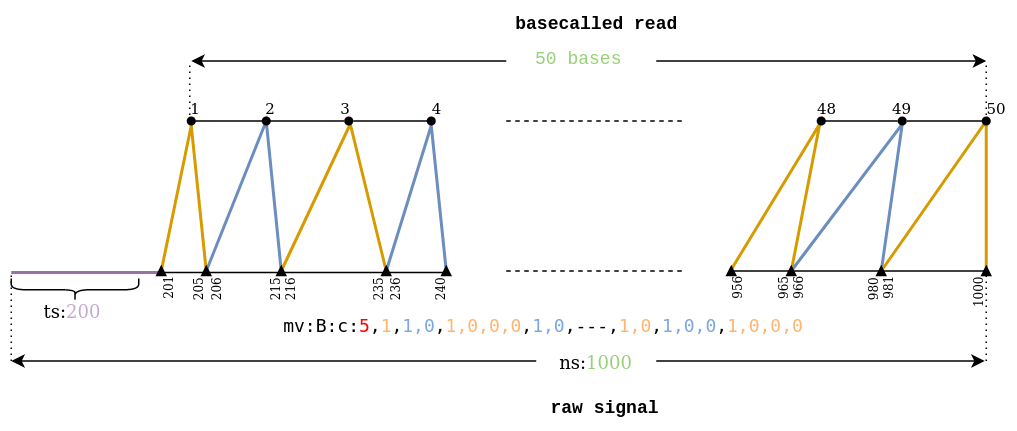
How the auxiliary field is stored in SAM format -> mv:B:c:5,1,1,0,1,0,0,0,1,0,1,0,1,0,0,0,1,0,1,0,1,1,0,1,0,1,0,1,1,1,1,…
Stride (always the first integer) -> 5
The actual move array (the rest) -> 1,1,0,1,0,0,0,1,0,1,0,1,0,0,0,1,0,1,0,1,1,0,1,0,1,0,1,1,1,1,…
The number of ones (1) in the actual move array equals to the fastq sequence length.
According to the above example the first move corresponds with 1 x stride signal points.
The second move corresponds with 2 x stride signal points. The third with 4 x stride, the fourth with 2 x stride and so on (see illustration below).
Different models have different strides. Strides from some of the guppy models are listed below.
| Model | Stride | Guppy version |
|---|---|---|
| dna_r10.4.1_e8.2_400bps_fast.cfg | 5 | v6.3.7 |
| dna_r10.4.1_e8.2_400bps_hac.cfg | 5 | v6.3.7 |
| dna_r10.4.1_e8.2_400bps_sup.cfg | 5 | v6.3.7 |
| dna_r9.4.1_450bps_fast_prom.cfg | 5 | v6.3.7 |
| dna_r9.4.1_450bps_hac_prom.cfg | 5 | v6.3.7 |
| dna_r9.4.1_450bps_sup_prom.cfg | 5 | v6.3.7 |
| rna_r9.4.1_70bps_fast_prom.cfg | 12 | v6.3.7 |
| rna_r9.4.1_70bps_hac_prom.cfg | 10 | v6.3.7 |
| dna_r10.4.1_e8.2_400bps_fast.cfg | 5 | v6.5.7 |
| dna_r10.4.1_e8.2_400bps_fast_prom.cfg | 5 | v6.5.7 |
| dna_r10.4.1_e8.2_400bps_hac.cfg | 5 | v6.5.7 |
| dna_r10.4.1_e8.2_400bps_hac_prom.cfg | 5 | v6.5.7 |
| dna_r10.4.1_e8.2_400bps_sup.cfg | 5 | v6.5.7 |
| dna_r9.4.1_450bps_fast.cfg | 5 | v6.5.7 |
| dna_r9.4.1_450bps_fast_prom.cfg | 5 | v6.5.7 |
| dna_r9.4.1_450bps_hac.cfg | 5 | v6.5.7 |
| dna_r9.4.1_450bps_hac_prom.cfg | 5 | v6.5.7 |
| dna_r9.4.1_450bps_sup.cfg | 5 | v6.5.7 |
| dna_r9.4.1_450bps_sup_prom.cfg | 5 | v6.5.7 |
| rna_r9.4.1_70bps_fast_prom.cfg | 12 | v6.5.7 |
| rna_r9.4.1_70bps_fast.cfg | 12 | v6.5.7 |
| rna_r9.4.1_70bps_hac.cfg | 10 | v6.5.7 |
| rna_r9.4.1_70bps_hac_prom.cfg | 10 | v6.5.7 |
As of dorado_v0.4.0 stride is used in three stages.
- In the process of creating chunks to feed into the NN model (code).
- In NN calculations on GPU (have a look at
stridein theconfig.tomlfile inside a model directory). - To properly stitch the output of each chunk to generate the final fastq sequence (code). This confirms that the NN model is down sampling the raw signal.